About Me
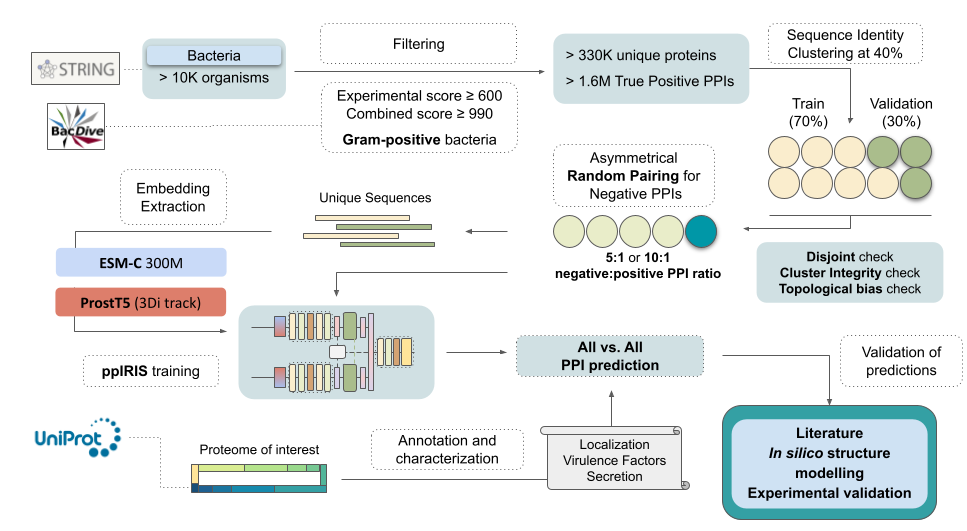
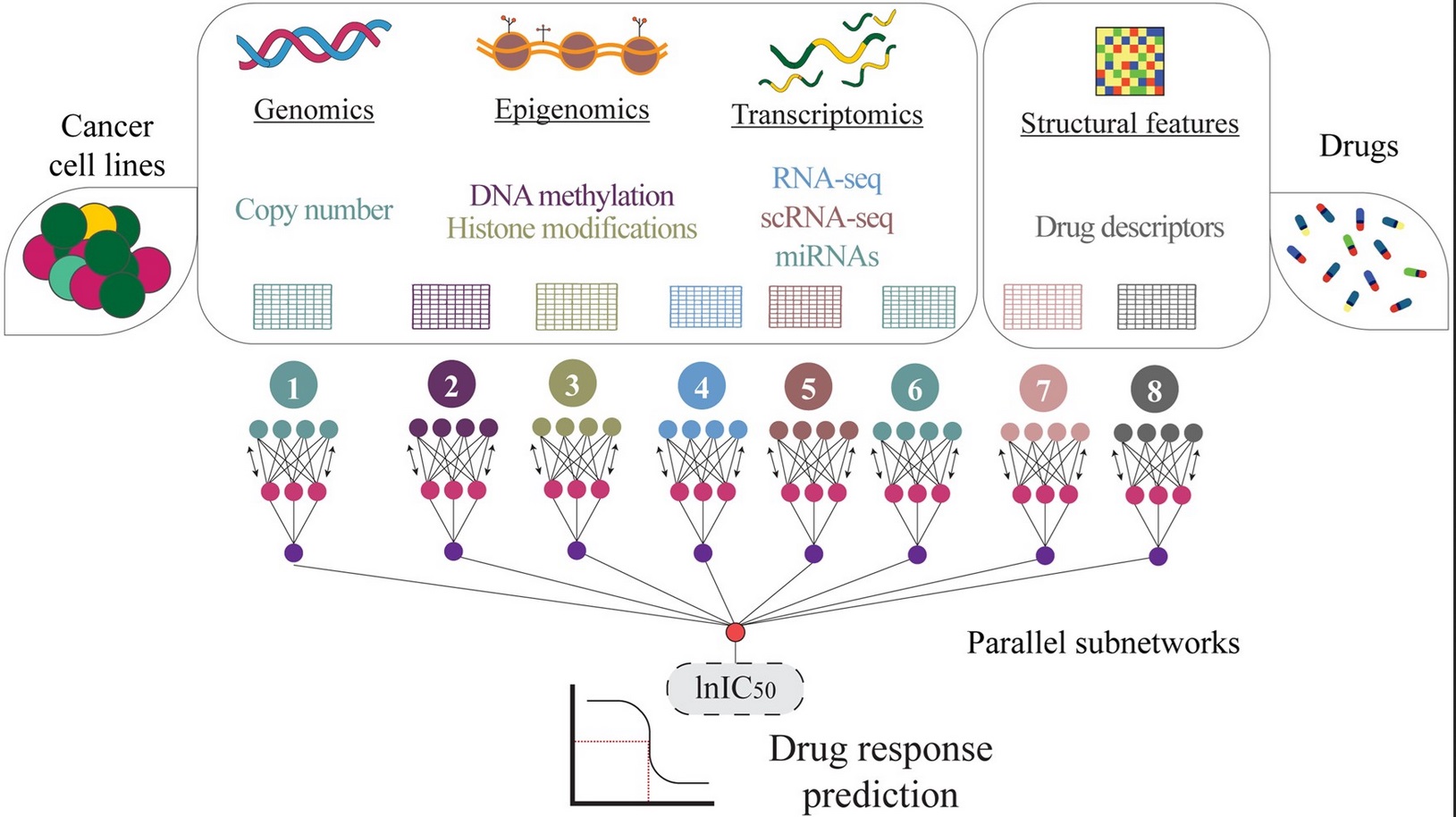
I'm a Computer Science PhD candidate at the National Institute for Research in Computer Science and Automation (INRIA). I really like using machine learning and deep learning to solve complex biological problems. Currently, I work mainly with the characterization of protein interactions and therapeutic protein design (from peptide binders to vaccines).
I believe my background in both life sciences and computer science allows me to approach research questions from a unique perspective, even in problems that do not directly involve biological data. After all, understanding biochemistry is to understand complex systems, which mirrors the intricacies of computational models.